Vignette: Paired total copy number analysis
Author: Henrik Bengtsson
Created: 2007-09-30
Last updated: 2008-10-27
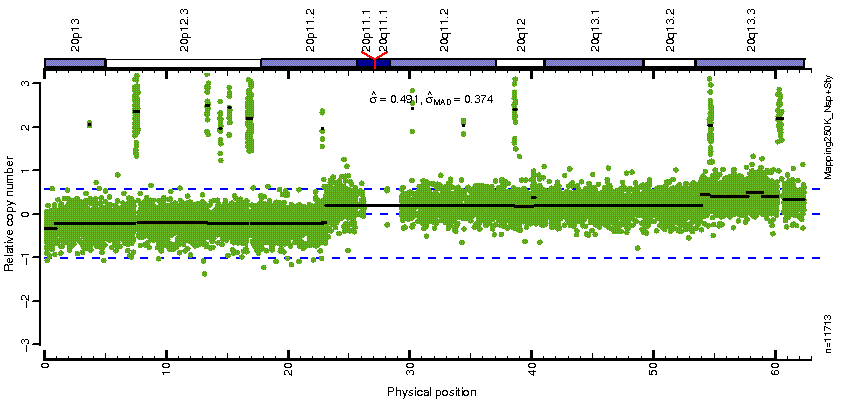
Figure: Raw copy-number estimates and CBS-region estimates on Chromosome 20 for tumor-normal pair (CRL-5868D, CRL-5957D).
To help support this work, please consider citing the following relevant references in your publications or talks whenever using their methods or results:
-
H. Bengtsson, K. Simpson, J. Bullard, et al. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Tech. rep. 745. Department of Statistics, University of California, Berkeley, Feb. 2008.
-
H. Bengtsson, R. Irizarry, B. Carvalho, et al. “Estimation and assessment of raw copy numbers at the single locus level”. Eng. In: Bioinformatics (Oxford, England) 24.6 (Mar. 2008), pp. 759-67. ISSN: 1367-4811. DOI: 10.1093/bioinformatics/btn016. PMID: 18204055.
Setup
Raw data
Data set: Affymetrix_2006-TumorNormal
Tumor-normal pairs: (CRL-2324D, CRL-2325D); (CRL-5868D, CRL-5957D); (CCL-256D, CCL-256.1D); (CRL-2320D, CRL-2319D); (CRL-2321D, CRL-2362D); (CRL-2336D, CRL-2337D); (CRL-2338D, CRL-2339D); (CRL-2340D, CRL-2341D); (CRL-2314D, CRL-2346D)
Chip types: Mapping250K_Nsp & Mapping250K_Sty
Source: This data set is publicly available. Look for public data sets on Page Mapping250K_Nsp & Mapping250K_Sty.
Note: The files downloaded from Affymetrix have 'NSP' and '_STY' parts of their filenames. You have to rename the files by replacing the '' (underscores) with ',' (commas), e.g. CRL-2324D_NSP.CEL to CRL-2324D,NSP.CEL and CRL-2324D_STY.CEL to CRL-2324D,STY.CEL. This way CEL files for the same sample gets the same name (the part before the first comma) so they can be automatically paired, e.g. CRL-2324D.
This is what the rawData/ directory should look like before starting the analysis:
rawData/
Affymetrix_2006-TumorNormal/
Mapping250K_Nsp/
CRL-2325D,NSP.CEL
...
CRL-5957D,NSP.CEL
Mapping250K_Sty/
CRL-2325D,STY.CEL
...
CRL-5957D,STY.CEL
Annotation data
If not already done, you also need to setup the annotationData/ directory for the two chip types. Please follow the instructions exactly as given in Vignette Total copy number analysis (10K, 100K, 500K).
Low-level analysis
library("aroma.affymetrix")
log <- Arguments$getVerbose(-4, timestamp=TRUE)
dataSetName <- "Affymetrix_2006-TumorNormal"
chipTypes <- c("Mapping250K_Nsp", "Mapping250K_Sty")
pairs <- matrix(c(
"CRL-2325D", "CRL-2324D",
"CRL-5957D", "CRL-5868D",
"CCL-256.1D", "CCL-256D",
"CRL-2319D", "CRL-2320D",
"CRL-2362D", "CRL-2321D",
"CRL-2337D", "CRL-2336D",
"CRL-2339D", "CRL-2338D",
"CRL-2341D", "CRL-2340D",
"CRL-2346D", "CRL-2314D"
), ncol=2, byrow=TRUE)
colnames(pairs) <- c("normal", "tumor")
Defining CEL set
csRawList <- list()
for (chipType in chipTypes) {
cs <- AffymetrixCelSet$byName(dataSetName, chipType=chipType)
stopifnot(all(getNames(cs) %in% pairs))
csRawList[[chipType]] <- cs
}
print(csRawList)
outputs
$Mapping250K_Nsp
AffymetrixCelSet:
Name: Affymetrix_2006-TumorNormal
Tags:
Path: rawData/Affymetrix_2006-TumorNormal/Mapping250K_Nsp
Chip type: Mapping250K_Nsp
Number of arrays: 18
Names: CCL-256.1D, CCL-256D, ..., CRL-5957D
Time period: 2006-01-13 13:07:38 -- 2006-01-16 10:18:56
Total file size: 1127.84MB
RAM: 0.02MB
$Mapping250K_Sty
AffymetrixCelSet:
Name: Affymetrix_2006-TumorNormal
Tags:
Path: rawData/Affymetrix_2006-TumorNormal/Mapping250K_Sty
Chip type: Mapping250K_Sty
Number of arrays: 18
Names: CCL-256.1D, CCL-256D, ..., CRL-5957D
Time period: 2006-01-18 12:57:07 -- 2006-01-18 20:30:57
Total file size: 1127.79MB
RAM: 0.02MB
Note how the two CEL sets have arrays with matching names, which is what aroma.affymetrix uses to pair up CEL files from the same sample. Note that paired CEL files still can have different tags, i.e. the filenames may differ as long as the part of the filenames that define the name is the same for both files. This is illustrated by the following code that give details for the first array in each set:
print(csRawList[["Mapping250K_Nsp"]][[1]])
AffymetrixCelFile:
Name: **CCL-256.1D**
Tags: **NSP**
Pathname:
rawData/Affymetrix_2006-TumorNormal/Mapping250K_Nsp/**CCL-256.1D,NSP.CEL**
File size: 62.66MB
RAM: 0.01MB
File format: v4 (binary; XDA)
Chip type: Mapping250K_Nsp
Timestamp: 2006-01-16 09:48:51
print(csRawList[["Mapping250K_Sty"]][[1]])
AffymetrixCelFile:
Name: **CCL-256.1D**
Tags: **STY**
Pathname:
rawData/Affymetrix_2006-TumorNormal/Mapping250K_Sty/**CCL-256.1D,STY.CEL**
File size: 62.65MB
RAM: 0.01MB
File format: v4 (binary; XDA)
Chip type: Mapping250K_Sty
Timestamp: 2006-01-18 14:59:36
Calibration for allelic crosstalk
csAccList <- list()
for (chipType in names(csRawList)) {
cs <- csRawList[[chipType]]
acc <- AllelicCrosstalkCalibration(cs)
print(acc)
csAcc <- process(acc, verbose=log)
csAccList[[chipType]] <- csAcc
}
Summarization
cesList <- list()
for (chipType in names(csAccList)) {
cs <- csAccList[[chipType]]
plm <- RmaCnPlm(cs, mergeStrands=TRUE, combineAlleles=TRUE, shift=+300)
print(plm)
fit(plm, verbose=log)
ces <- getChipEffectSet(plm)
cesList[[chipType]] <- ces
}
PCR fragment length normalization
cesFlnList <- list()
for (chipType in names(cesList)) {
ces <- cesList[[chipType]]
fln <- FragmentLengthNormalization(ces)
print(fln)
cesFln <- process(fln, verbose=log)
cesFlnList[[chipType]] <- cesFln
}
This gives raw CN estimates for the 18 samples for both chip types.
Identification of copy-number regions in tumor-normal pairs
# Split data set in (tumor, normal) pairs
sets <- list(tumor=list(), normal=list())
for (chipType in names(cesFlnList)) {
ces <- cesFlnList[[chipType]]
for (type in colnames(pairs)) {
idxs <- match(pairs[,type], getNames(ces))
sets[[type]][[chipType]] <- ces[idxs]
}
}
cns <- CbsModel(sets$tumor, sets$normal)
print(cns)
# Link the ChromosomeExplorer to the segmentation model
ce <- ChromosomeExplorer(cns)
print(ce)
# Fit the model for a few chromosomes
process(ce, chromosomes=c(1, 19, 22), verbose=log);
# The X chromosome is very noisy and generates quite a few missing values
process(ce, chromosomes=23, maxNAFraction=1/5, verbose=log)